RNA polymerase 3D of poliovirus
Poliovirus carries the information for it's own RNA polymerase (RdRP, RNA-dependent RNA Polymerase). The structure of this enzyme resembles that of most other nucleic acid polymerases, which reminds you of a half-opened hand. So you find domains called palm
connected to a strong thumb
and a finger domain
. Unique to the poliovirus polymerase are aminoterminal structures not seen in other polymerases
. The structurally related domains are functionally related too, with the fingers and thumb involved in substrate binding (primer and template) and the palm involved in phosphoryl transfer.
Note several loose ends in this model
- there are regions with insufficiently dense electron patterns in the X-ray analysis to exactly locate amino acid atoms, which are consequently omitted here (always use the mouse to turn the molecule at will for different aspects - you may notice then the now hidden parts!).
The palm domain is composed of motifs which are found in most nucleic acid polymerases as well.
Met225 - Leu241At the end of the short helix of motif A you find the structurally conserved Ser240 . It hydrogen bonds to carbonyl oxygens of both Gly236 and Tyr237 and so stabilizes this helical loop.
Cys290 - Thr312
Leu318 - His336
Asp339 - Met354
Glu369 - Ala380
Within motif A there also is a completely conserved aspartate - in all polymerases this amino acid coordinates a catalytically neccessary metal ion .
Another set of amino acids is conserved in RNA-dependent RNA-polymerases: Asp238 in motif A and Asn297 in motif B . At these very positions aromatic amino acids are found in RNA-dependent DNA-polymerases (Tyr and Phe in HIV reverse transcriptase). So these residues are thought to discriminate betweeen ribo- and desoxyribonucleotides in the polymerization process.
The beta-turn in motif C harbours more catalytically important aspartates . Mutation of Asp233 or Asp328 will inactivate the polymerase, a change of Asp329 to Asn alters the metal specificity .
Motif E is unique to RNA-dependent polymerases. Amino acids in this motif interact with the nascent primer strand in the polymerization process. Structurally it connects to the thumb domain by forming a sheet .
In this model of polio RdRP a large part of the finger domain is not visible . However, you might have an impression of the shape of the whole enzyme by comparing it with a structurally highly homologous RdRP from rabbit hemorrhagic disease virus (which is 40 amino acids larger than the polio enzyme) . In spacefilling view you notice the 'hand' to be completely closed with just a hole to pass the substrate RNA through. The known part of the polio RdRP occupies the same volume, with the exception of the aminoterminal part (residues 12 - 24) which obstructs the channel in the enzyme. This may be due to a crystallisation artefact, as no RNA was present in the experiment.
Highly purified polio RdRP is capable of primed RNA synthesis on its own in vitro. In cellular environments the enzyme oligomerizes into complex lattices which are associated with membraneous structures. Amino acids forming two interfaces at cross angles were identified in the molecule. Interface I is formed by 23 amino acids at opposite sides of the structure, enabling (theoretically unlimited) threads of head-to-tail arrangements. The other interface (II) will allow for another threading giving rise to a twodimensional lattice. A metal ion is involved here. The proximity of polymerase molecules in this arrangement gives rise to the efficiency of coupled plus and minus strand synthesis or recombinational events.
3D polymerase of polio is in itself not a membrane bound protein. Fastening to membranes is mediated by another viral protein, 3AB. Distinct amino acids of polio polymerase are involved in binding to 3AB which are situated perpendicular to the plane of the lattice forming interfaces .
To give you an impression of the intracellular working place of the polymerase, an electron micrograph of an isolated replication complex is shown here. This distinct structure of membraneous lobes is called a rosette.
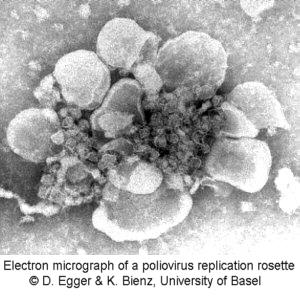
this demonstration.
Literature:
JL Hansen et al, Structure of the RNA-dependent RNA polymerase of poliovirus, Structure 5 (1997) 1109-1122
DW Gohara et al, Poliovirus RNA-dependent RNA polymerase, J. Biol. Chem. 275 (2000) 25523-25532
HB Pathak et al, Structure-function relationships of the RNA-dependent RNA polymerase from poliovirus (3Dpol), J. Biol. Chem. 277 (2002) 31551-31562
JM Lyle et al, Visualization and functional analysis of RNA-dependent RNA polymerase lattices, Science 296 (2002) 2218-2222
D Egger & K Bienz, Recombination of poliovirus RNA proceeds in mixed replication complexes originating from distinct replication start sites, J. Virol. 76 (2002) 10960-10971
KKS Ng et al, Crystal structures of active and inactive conformations of a calciviral RNA-dependent RNA polymerase, J. Biol. Chem. 277 (2002) 1381-1387

