Protomer of poliovirus capsid
The PDB structure file for this script is derived from 1hxs.unique; it had to be adapted so that Chime/Jsmol is capable to handle it. In compressed form it is still 1 MB in size, so please be patient until the data are loaded.
The capsid of poliovirus is made up of 60 subunits which contain four protein chains each. Shown here is one subunit (or protomer) containing:
-
VP1
(302 amino acids)
-
VP2
(272 amino acids)
-
VP3
(238 amino acids)
-
VP4
(68 amino acids)
VP4 is synthesized along with VP2 and cleaved off only when the complex (VP0) is inserted into the capsid. The PDB entry notes the myristyl residue at the amino terminus of VP4 as #1 .
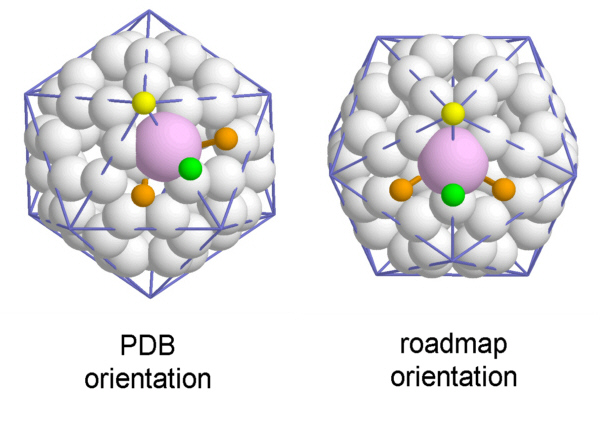
The choice of which viral proteins constitute a protomer is somewhat arbitrary and made here in analogy to plant viruses with similar structure. The atomic structure data are deposited in the PDB in the configuration shown to the left : One 5-fold axis is placed north and one 3-fold Axis to the south. However, you might as well choose the 'roadmap'-view by selecting the right hand side VP3 :
The folding pattern in the three major proteins is rather the same: there is a large beta-sheet composed of eight antiparallel strands which is wrapped into a distorted barrel. When unwrapped into a plane, the sheets clearly show a jelly-roll structure (also termed Greek key motif).
VP1 beta sheet
VP1 helices
VP2
protein chain
VP2
beta sheet
VP2
helices
VP3 beta sheet
VP3 helices
Upon closer inspection you may notice the beta barrels are not completely closed, so the term beta sandwich may be more appropriate.
VP4
protein chain
VP4
sheet (with VP1)
VP4
helices
The structures of VP1 to VP3 can be abstracted to a common pattern. In the literature you find the following strand and loop designations:
As may be guessed from the entangeled trace of the Calpha-atoms there are tight contacts between the proteins constituting a protomer. It becomes evident when looking at the proteins in spacefill mode . The gaps at the edges of the protomer are filled by neighboring subunits in the assembled capsid, as will be shown in the next section.
Interactions of protomers
The data used here to show the protein model were gained from whole virions, they do not represent the structure of a single protomer in solution. So the structure has to be considered in connection with the other protomers. Interactions are of special interest around the symmetry axes of the virus.
A fivefold axis
is surrounded on the surface of the particle by VP1. VP4 is completely covered by the other proteins, it is accessible only from inside
. The aminoterminal part of VP4 contacts several other protomers around the axis
. Also, in the pentamer there are additional structural elements pertaining to it's stability
. The amino termini of VP3 find together to form a beta barrel
. Additional short beta-sheets are formed on the inner surface by stretches of VP1 and two VP4 (from the same and an adjoining protomer)
. The myristoyl residues of VP4 create a kind of hydrophobic plug on the inside below the beta barrel
.
The loops of VP1 near to the fivefold axis form a turret protruding from the surface
. The loops contain charged amino acids generating electrostatic forces
as well as hydrophobic residues
to glue the protomers together by van der Waals forces. In a cutaway view of the pentamer there is a deep groove to be seen beneath the VP1-turret
which you find in the literature termed canyon.
A view down a threefold axis shows alternately VP2 and VP3 on the outer surface. Inside all VPs are seen, penetrating each other. One special feature makes the protomers stick together in the interfaces around this axis. One part of the beta sandwich in VP3 is extended by the 'extra' sheet of VP2 (residues 14 - 25) of the neighboring protomer . This sheet is even further enlarged by amino acids of VP1 of the first protomer , so the protomers 'plug in' to each other (which again is reason enough to open the VP3 barrel into a sandwich). This feature is found in all three interfaces around the axes, of course .
Protomer interactions around the twofold axes are defined by the interactions around the threefold axes . Additionally, there are helices in VP2 facing each other at the very position of the twofold axes. Their amino acids are in close contact .
this demonstration.
Literature:
JM Hogle et al, Three-dimensional structure of poliovirus at 2.9 Å resolution, Science 229 (1985) 1358-1365
ST Miller et al, Ab initio phasing of high-symmetry macromolecular complexes: successful phasing of authentic poliovirus data to 3.0 Å resolution, J. Mol. Biol. 307 (2001) 499-512
C Zhang & SH Kim, A comprehensive analysis of the Greek key motifs in protein beta-barrels and beta-sandwiches, Proteins Struct. Funct. Genet. 40 (2000) 409-419

