Spike protein of corona viruses
Here you see a model of the spike protein of sars-cov2. The atoms comprising the amino acids are shown as green spheres, the chocolate brown parts are attached sugar molecules.
The spike protein is anchored to the viral membrane with it's "lower" part (to visualize click
, anchor sequence red).
The "upper" part of the protein binds to the receptor, angiotensin converting enzyme 2 (RBD = receptor binding domain colored yellow)
. Within this domain is the receptor binding motiv
. In the close up
you find the amino acids with direct contact to the receptor
.
The spike protein is not on its own - on the viral surface it is always arranged as trimers intertwined around a threefold symmetry axis . The receptor binding domain of the spike protein may assume different positions. In the model investigated here one RBD is in the "up" position, the other two adopt the "down" position . The interacting surface of the RBM part are "asleep" in the "down" position.
Spike trimers are abundant on the virus' surface - with more infectious variants even more so, compared to the "harmless" cold-only-variants of the corona species.
this demonstration.
Literature:
D. Wrapp et al, Science 367, 1260-1263 (2020), DOI: 10.1126/science.abb2507
J Lan et al, Nature 581, 215-220 (2020), DOI: 10.1038/s41586-020-2180-5
Another group of scientists made a movie of the "down" to "up" movement of the RBD by interpolating the atomic coordinates:
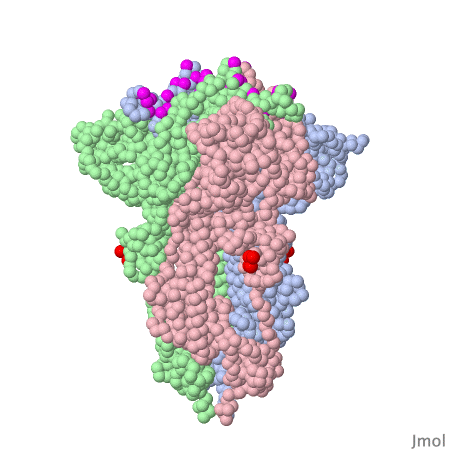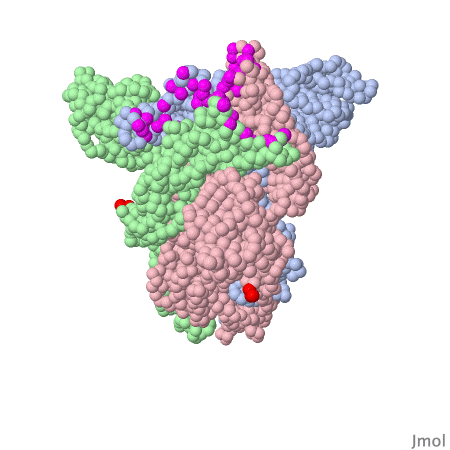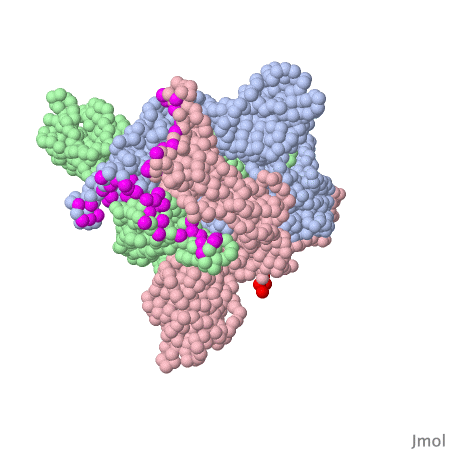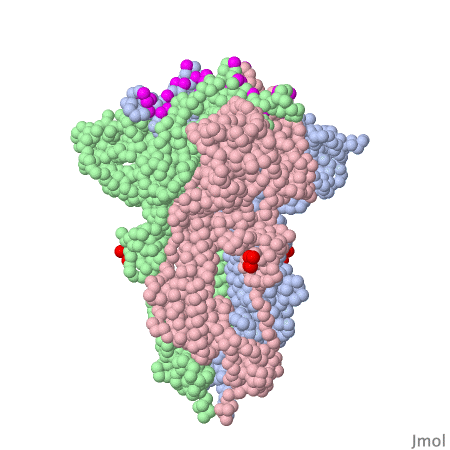
Amino acids contacting the ACE2 receptor are marked violet.
Reference:
Protopedia, https://proteopedia.org/wiki/index.php/SARS-CoV-2_protein_S_priming_by_furin
A G Wrobel et al, Nat Struct Mol Biol 27, 763-767 (2020), DOI: 10.1038/s41594-020-0468-7

