Traditional Chinese Medicine - with chinese computing power
Treatment of diseases with natural substances (plant parts or extracts) was practised for thousends of years all over the world before the invention of modern pharmaceutical products. Herbal drugs are still widely administered, based on experience for lack of detailed knowledge about active ingredients. Physicians nowadays like to know the mechanism of prescribed drugs. So chinese scientist made an effort to collect all known facts about traditionally used plants and their constituents; the result is the Traditional Chinese Medicine System Pharmacology Database (TCMSP), which is publicly accessible (https://tcmsp-e.com/). The database offers both chinese and english front ends.
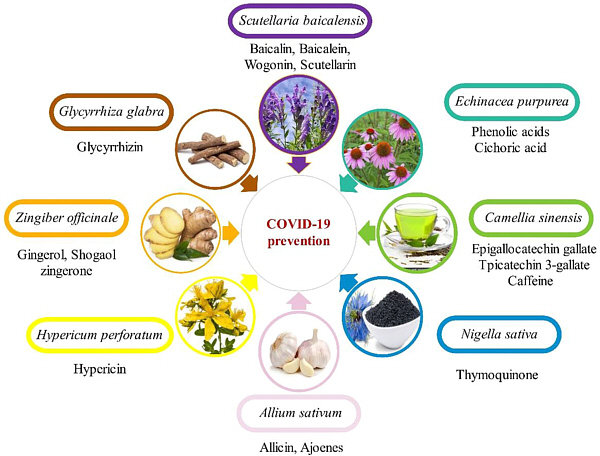 Herbal drugs were found to have an effect on covid-19 by modulating the immune system or interfering directly with virions. Boozari & Hosseinzadeh describe some plants and speculate about mechanisms of action (see figure for selected plants).
Herbal drugs were found to have an effect on covid-19 by modulating the immune system or interfering directly with virions. Boozari & Hosseinzadeh describe some plants and speculate about mechanisms of action (see figure for selected plants).
More recently Wang et al published a survey of 65 ingredients of natural substances with known in vitro inhibitory effects against corona virus infections.
Bioinformatics to search for drugs
For a really efficient substance to cure SARS its properties have to be known, its target, its interaction mechanism, and a means to extract and enrich it. At the time of this writing the TCSMP contains data for more than 12.000 chemical substances, including their atomic structures. Which ones might be promising to start experimental research (with the aim to get results within a lifetime)? Luckyly we live in the time of supercomputers and all kinds of modelling and docking simulation programs. With the aid of genetic and protein property databanks substances of the TCSMP may be matched to target structures and their molecular interactions in silico investigated. In the first run this requires no hands on laboratory work, you just have to pay for the computing time.
Finding active ingredients of licorice
Reducing a possible 12.000 substances to 11 promising ones for treatment of covid2 was achieved by preselecting ingredients of the roots of Glycyrrhiza glabra (commonly known as licorice) for the in silico investigation.
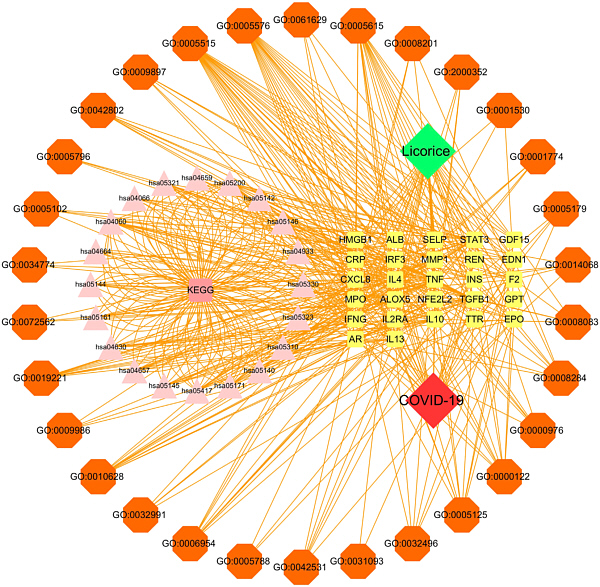 For this investigation Cao et al at first used the GeneCard human genes database to find targets of both licorice compounds and Cov-2. 101 combined targets were found. Using the databanks GeneOntology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) resulted in a complex network of interdependencies. You needn't understand the details of the figure, it is just presented to give an impression of the found interdependencies. More calculations to estimate the interaction strength found 11 licorice ingredients to bind tightly to their covid-related targets. For each of these complexes the molecular details of the binding were evaluated by virtual molecular docking in silico. Eventually the prediction of binding energies of the ligands led to the three strongest candidates for covid-19 treatment. Now the predicted effects - influencing the immune response and regulate cell survival - have to be verified by experiments in living systems.
For this investigation Cao et al at first used the GeneCard human genes database to find targets of both licorice compounds and Cov-2. 101 combined targets were found. Using the databanks GeneOntology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) resulted in a complex network of interdependencies. You needn't understand the details of the figure, it is just presented to give an impression of the found interdependencies. More calculations to estimate the interaction strength found 11 licorice ingredients to bind tightly to their covid-related targets. For each of these complexes the molecular details of the binding were evaluated by virtual molecular docking in silico. Eventually the prediction of binding energies of the ligands led to the three strongest candidates for covid-19 treatment. Now the predicted effects - influencing the immune response and regulate cell survival - have to be verified by experiments in living systems.
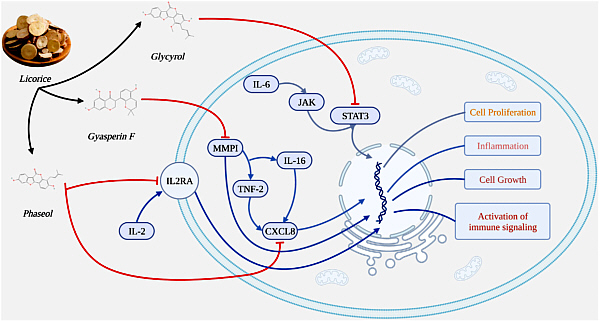 The summarizing figure shows the three substances acting on cellular components.
The summarizing figure shows the three substances acting on cellular components.
Rotating to the left are the molecular structures of (from the top)
Glyasperin F
Glycyrol
Phaseol
References:
M Boozari & H Hosseinzadeh, Natural products for COVID-19 prevention and treatment regarding to previous coronavirus infections and novel studies, Phytotherapy Research 2021;35:864-876
Z Wang et al, Bioactive natural products in COVID-19 therapy, Front. Pharmacol. 2022;13:926507. doi: 10.3389/fphar.2022.926507
JF Cao et al, Exploring the mechanism of action of licorice in the treatment of COVID-19 through bioinformatics analysis and molecular dynamics simulation, Front. Pharmacol. 2022; 13:1003310. doi: 10.3389/fphar.2022.1003310

