| UHH > Faculty of Biology > Teaching Stuff > Highlights of Biochemistry > Photochemistry | |
Bacterial Photosynthesis
Overview
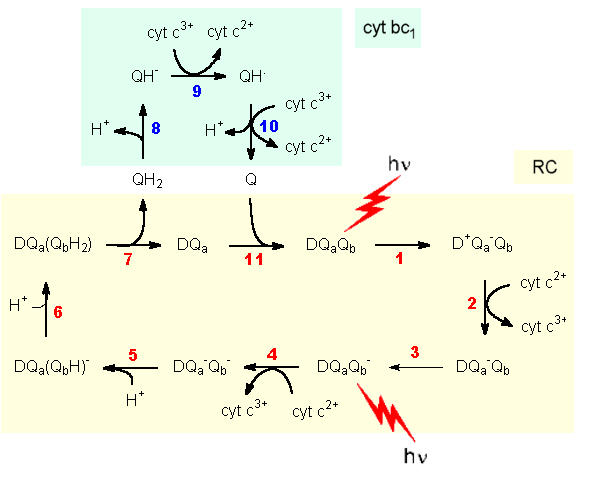 The initial step of the energy conversion sequence is the electronic excitation of a chlorophyll in the reaction center upon receiving a photon's energy. The state of the chlorophyll is relaxed by transferring an electron to a quinone, defining the chlorophyll as the primary donor. In step 1 thus a charge couple D+Q- is generated. In step 2 the chlorophyll is reduced by a cytochrome, and in step 3 the charge of the quinone is passed on to a second quinone. Step 4 repeats steps 1 and 2 resulting in two charged quinones. In steps 5 and 6 protons are taken up to generate an uncharged quinone and a hydroquinone. The hydroquinone diffunds out of the reaction center into an also membrane bound cytochrome bc1 complex and is reoxidized to the quinone state. In steps 8 and 10 protons are liberated. Due to the spatial arrangement of the protein complexes the protons are taken up in steps 5, 6 in the interior of the cell and expelled in steps 8, 10 to the periplasmic space. This process generates an electrochemical gradient across the cytoplasmic membrane. The quinone cycle is closed by transfer of the quinone back to the RC (11).
The initial step of the energy conversion sequence is the electronic excitation of a chlorophyll in the reaction center upon receiving a photon's energy. The state of the chlorophyll is relaxed by transferring an electron to a quinone, defining the chlorophyll as the primary donor. In step 1 thus a charge couple D+Q- is generated. In step 2 the chlorophyll is reduced by a cytochrome, and in step 3 the charge of the quinone is passed on to a second quinone. Step 4 repeats steps 1 and 2 resulting in two charged quinones. In steps 5 and 6 protons are taken up to generate an uncharged quinone and a hydroquinone. The hydroquinone diffunds out of the reaction center into an also membrane bound cytochrome bc1 complex and is reoxidized to the quinone state. In steps 8 and 10 protons are liberated. Due to the spatial arrangement of the protein complexes the protons are taken up in steps 5, 6 in the interior of the cell and expelled in steps 8, 10 to the periplasmic space. This process generates an electrochemical gradient across the cytoplasmic membrane. The quinone cycle is closed by transfer of the quinone back to the RC (11).
In the frame to the right the chromophores of the reaction center of Rhodopseudomonas sphaeroides are to be seen. The primary chlorophyll (type a) is shown in yellow. The electrons are passed via another bacteriochlorophyll a (green) and a bacteriopheophytin (light blue) to ubiquinone-10 (red). Magnesium atoms are symbolized in dark green, a single iron ion in orange.
The primary chlorophyll in the reaction center is rather a small target to be hit by photons. To improve the absorption cross section, RCs are surrounded by antenna proteins harbouring more chlorophyll as well as pigments absorbing at other wavelengths thus extending the efficiency even further. These proteins are arranged in circular patterns around the RCs within the bacterial cytoplasmic membrane. The diameter of these complexes extends to ca. 10 nm, they are termed light harvesting complex I (LH I). Most bacteria contain additional photosensitive complexes (LH II). An example of the chromophore arrangement of a LH II is shown at the top of this page. Below two different complexes are shown with their additional carotenoid chromophores and the supporting protein structure. In the bacterial membranes several LH II complexes are situated close enough to LH I to mediate fast energy transfer via LH I to the RC.
Nonameric LH II from Rhodopseudomonas acidophila
Protein - Chlorophyll a - Rhodopin-glucoside
Octameric LH II from Rhodospirillum molischianum
Protein - Chlorophyll a - Lycopene
Reaction center from Rhodopseudomonas viridis
Protein chain L - chain M - chain H - Cytochrome chain C - Chlorophyll b - Pheophytin b - Quinone - Heme - Iron - Magnesium
![]()
The core proteins of the RCs of purple bacteria are the L and M chains which give the complex a quasi symmetric structure. They span the membrane with five helices each and harbour the active ligands. In most of these bacteria an asymmetrically placed H chain is found which also spans the membrane with one helix. Only in some species a cytochrome c chain is integrated into the RC.
Despite the nearly symmetrical arrangement of the chromophores there is a functional difference: the a branch is 200 times more used in electron transport than the b branch.
Within the membranes RCs and cytochrome bc1 complexes are found in a stochiometry of 2:1. Electron micrographs of native membranes containing two-dimensionally ordered PSUs from LH I, RC and cyt bc1 led to a model showing two RCs placed symmetrically beneath a cyt bc1 surrounded by c-shaped LH I-complexes comprised of 12 subunits in Rhodobacter sphaeroides.
Literature:
MY Okamura & G Feher, Proton transfer in reaction centers from photosynthetic bacteria, Ann. Rev. Biochem. 61 (1992) 861-896
RE Blankenship, http://photoscience.la.asu.edu/photosyn/education/antenna.html
CRD Lancaster & H Michel, The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB, Structure 5 (1997) 1339-1359
S Iwata et al, Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex, Science 281 (1998) 64-71
C Jungas et al, Supramolecular organization of the photosynthetic apparatus of Rhodobacter sphaeroides, EMBO J. 18 (1999) 534-542
AW Roszak et al, Crystal structure of the RC-LH1 core complex from Rhodobacter palustris, Science 302 (2003) 1969-1972
About the basics of membrane proteinology:
G v Heijne, A day in the life of Dr. K. or how I learned to stop worrying and love lysozyme: A tragedy in six acts, J. Mol. Biol. 293 (1999) 367-379
2-2000 © Rolf Bergmann